Single-Stranded DNA for Improved Knockin Efficiency: Animal Model Generation
The CRISPR/Cas9 gene-editing tool kit has facilitated gene knockins above any other strategy developed to date. Still, improving efficiency and specificity remain challenging aspects of CRISPR/Cas9 directed gene insertions. In a continued effort to enhance knockin efficiency, investigators have relied on different approaches, including optimizing guide RNAs, increasing HDR frequency, improving co-localization of Cas9 and donor templates to targets sites, and choosing the right donor template.
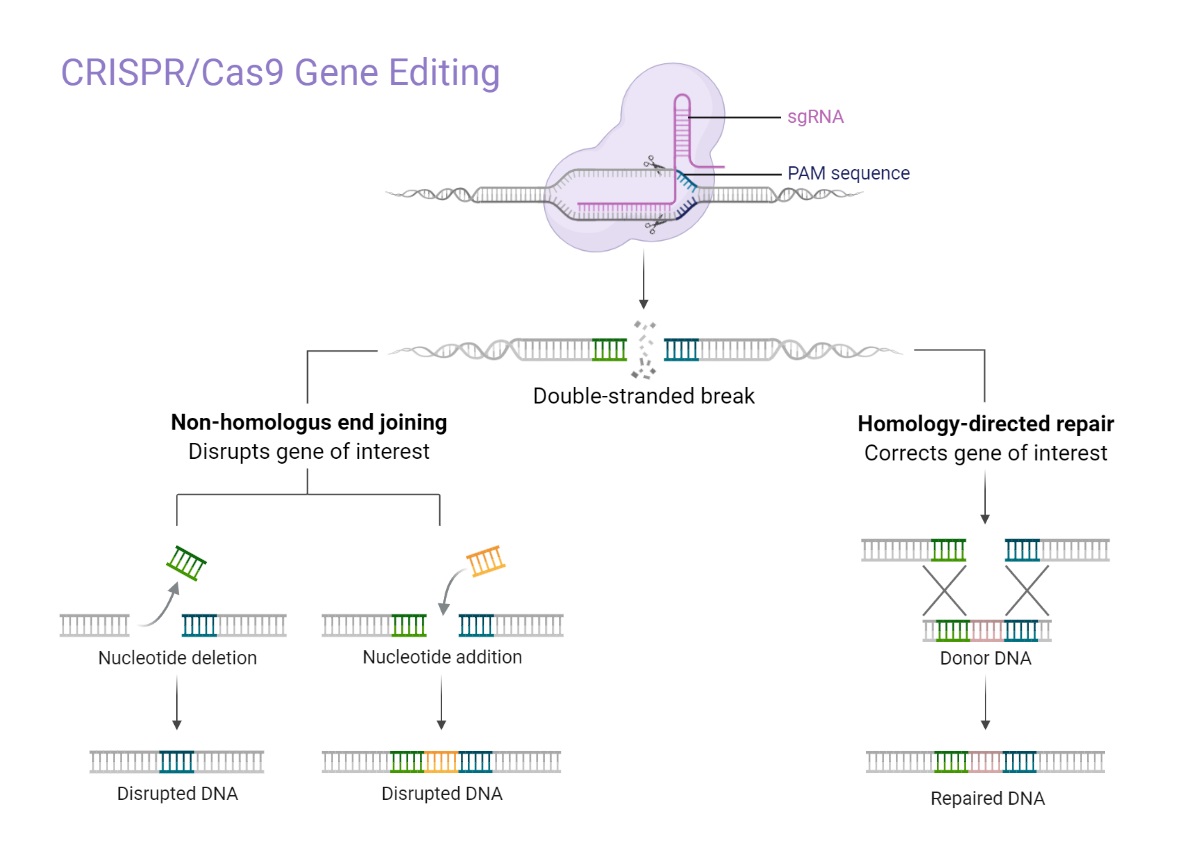
Reprinted from "CRISPR/Cas9 Gene Editing", by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
CRISPR/Cas9 complex binds and cuts a target DNA upstream of a PAM sequence. DNA double-strand breaks lead to the activation of two principal mechanisms for repair: Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR). NHEJ results in insertions or deletions (indels), and HDR leads to precise repairs. For insertions, nuclease based gene editing strategies depend on the activation of HDR. The low frequency of this pathway limits the efficiency of knockins.
When it comes to knockin approaches, not all donor templates are equally efficient. Several studies have achieved greater knockin efficiency across different animal models, including mouse, rat, and zebrafish, by using single-stranded DNA (ssDNA) donor templates as opposed to double-stranded DNA (dsDNA) (Yoshimi et al. 2016, Miura et al. 2018, Ranawakage et al. 2021).
In mice, the use of long single-stranded DNA (lssDNA) donor templates combined with the CRISPR ribonucleoprotein (CRISPR-RNP) complex led to substantially improved knockin efficiency. This strategy for the generation of mouse knockin models referred to as Easi-CRISPR, which stands for (Efficient additions with ssDNA inserts-CRISPR), has high insertion efficiencies ~30-60% (Miura et al. 2018).
Recently, Ranawakage et al. demonstrated a similarly improved insertion efficiency in zebrafish when relying on lssDNA and CRISPR-RNP complexes to target several genes, including sox3, sox11a, and pax6a. When using different DNA constructs, consisting of ~800 nucleotide lssDNA or dsDNA plasmid and PCR fragment for knockin of composite tags, Ranawakage et al. found the insertion efficiency of lssDNA (~90%) to outperform that of dsDNA (~75% PCR and ~15% plasmid). Moreover, based on embryo survival after 48hpf, lssDNA donor templates led to lower toxicity than dsDNA.
Additionally, Ranawakage and colleagues found that lssDNA donor template strand choice, namely target or non-target strand, influences insertion efficiency and needs to be optimized for each sequence and loci. For example, knockin efficiency at a sox3 site was increased using a target strand lssDNA donor template. Interestingly, knockin frequency at sox11a and pax6a target sites benefited from using a non-target strand lssDNA donor template. Lastly, they found that the length of the 3' homology arm needs optimization as insertion may be more efficient at some sites when designing lssDNA with a shorter homology arm (e.g., 50 vs. 300 nucleotides).
Overall, investigators have found several advantages to using ssDNA donor templates for CRISPR/Cas9 mediated knockins, including increased insertion efficiency and greater on-target integration (Quadros et al. 2017, Miyasaka et al. 2018). The higher cytotoxicity of dsDNA found by Ranawakage et al. agrees with other investigators' reports, suggesting that ssDNA donor templates may be a better choice for undertaking the time-consuming and laborious processes involved in knockin animal model generation (Roth et al. 2018).
Recently, GenScript collaborated with Dr. Brian Lin at Massachusetts General Hospital, Boston, MA, on his project, which used CRISPR/Cas9 ribonucleoprotein combined with lssDNA for generating a tetracycline-inducible mouse line, Foxi1-rtTA. Learn more Case Study.
- Like (2)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2026 MolecularCloud



