Driving Antibody Development through Improved Engineering Technologies
In recent years monoclonal antibodies have increasingly become sought after tools for therapy. These highly specific molecules, mostly relegated to basic research labs as tools for in vitro and in situ protein analysis, have emerged as strong candidates for therapy in a broad range of indications over the years. Currently, monoclonal antibody therapies have been approved or are being developed for cancer (e.g., multiple myeloma), infectious disease (e.g., COVID-19), neurodegeneration (e.g., Alzheimer's disease), autoimmune disease (e.g., psoriasis), and metabolic disease (e.g., high LDL bad cholesterol) (Lu et al. 2020).
Based on the Antibody Society, 12 therapeutic antibodies were approved by the FDA in 2020, and an additional 16 were under review and pending approval by the year's end. Driving this growing trend are various synthetic biology approaches, which have pushed the boundaries in discovering and developing antibodies with reduced adverse effects and improved efficacy. Advancements in antibody engineering technologies, including CDR grafting for monoclonal antibody humanization and phage display for developing fully human antibodies, progressively improved antibody drugs' discovery and development. Additionally, phage display technology, together with mutagenesis approaches, has supported optimizing antibody properties to achieve greater specificity, affinity, and function (Lu et al. 2020).
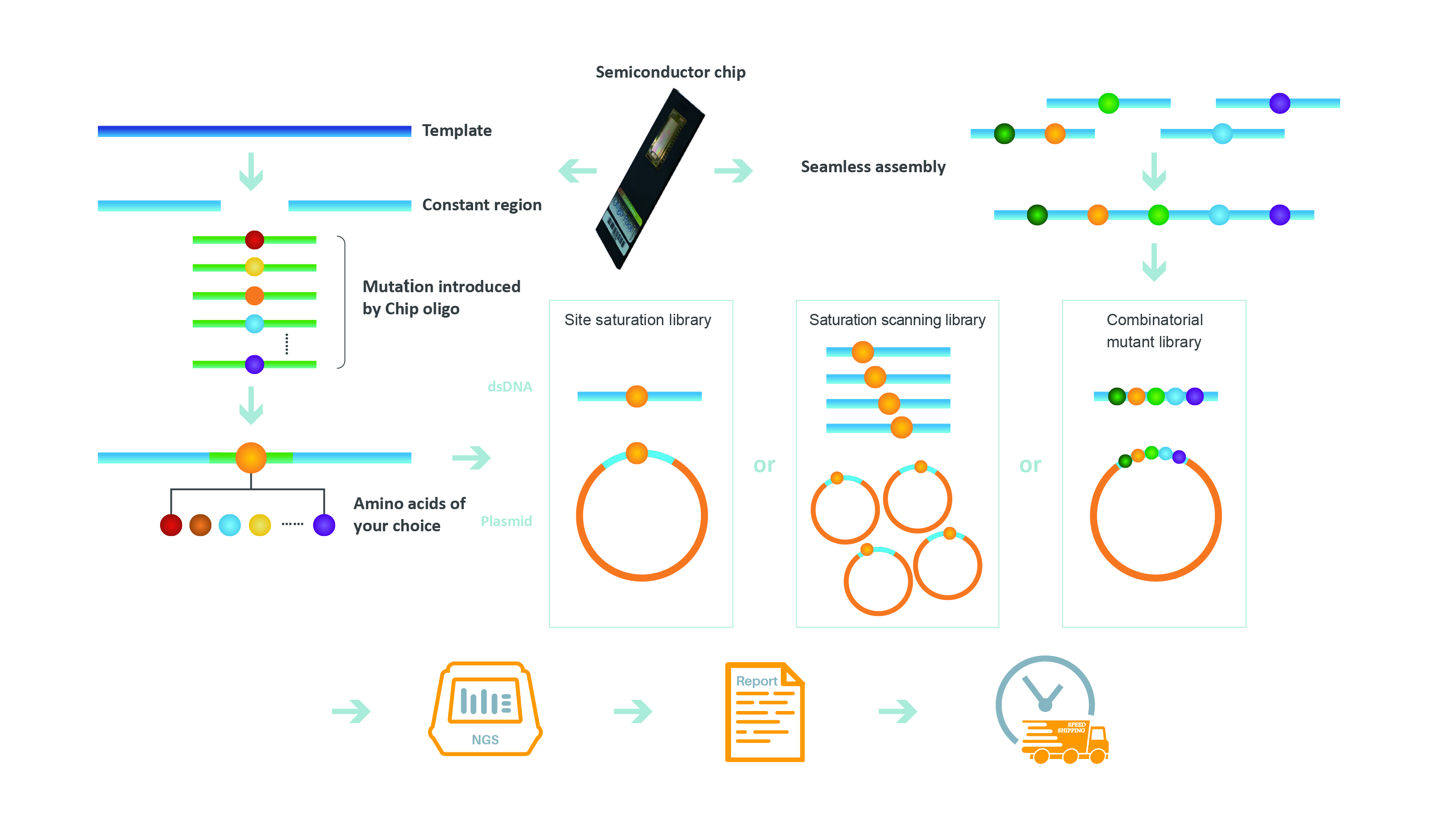
GenScript's Precision Mutant Libraries Platform Workflow. Oligonucleotides with specified mutations are generated by arrayed semiconductor-based DNA synthesis for incorporation into suitable vectors. Semiconductor-based oligonucleotide synthesis supports the design of three main types of libraries, including site-saturation, saturation scanning, and combinatorial mutant libraries. Library sequences are verified through NGS and made available to investigators as double stranded-DNA or cloned into plasmids. Learn more about Precision Mutant Libraries, Application Note.
Fine-tuning Antibody-drug Properties
Presently, most therapeutic antibodies are identified from large libraries (e.g., scFv and Fab-fragment libraries). These antibody libraries are subject to multiple selection cycles, ultimately identifying specific antigen-binders (Lu et al. 2020). Antibody leads are commonly modified further by various engineering strategies to improve desirable properties such as affinity.
Affinity maturation is a biological process that supports improved antibody-antigen binding and function. In vivo, this process occurs in germinal centers within lymphoid tissues and involves repeated hypermutation and selection cycles (Lim et al. 2019). However, this process may be replicated in vitro by constructing mutant libraries where changes in the sequence of antibodies, for example, within the CDR or antigen-binding region, are introduced randomly, semi-rationally, or rationally. Random mutagenesis may be achieved through error-prone PCR or by degenerate codons (Lim et al. 2019, Romero et al. 2009). Directed evolution or random mutagenesis is the method of choice when limited information is available relating sequence to antibody function.
In contrast, if the relation between antibody sequence, structure and function is known, a semi-rational (e.g., saturation mutagenesis) or rational (e.g., site-directed mutagenesis) library design approach may be implemented (Lim et al. 2019). A significant advantage of these more focused methods is the reduced size of the mutant libraries, decreasing screening efforts. Among semi-rational engineering strategies, precision mutant libraries are developed by leveraging the specificity of semiconductor-based DNA synthesis. In this platform, precise nucleotide changes are introduced, allowing complete control over codon usage, and resulting in mutant libraries with reduced bias and optimal codon distribution. GenScript scientists have developed an application note describing how arrayed semiconductor-based DNA synthesis enables generating a saturation mutant library targeting specific sites within an antibody's CDR regions for improved affinity in the femtomolar range.
Learn more, nature research "Controlling codes for enhanced proteins"
Reference
Lim, C. C., Choong, Y. S. & Lim, T. S. Cognizance of molecular methods for the generation of mutagenic phage display antibody libraries for affnity maturation. Int. J. Mol. Sci. (2019) doi:10.3390/ijms20081861.
Lu, R. M. et al. Development of therapeutic antibodies for the treatment of diseases. Journal of Biomedical Science (2020) doi:10.1186/s12929-019-0592-z.
Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nature Reviews Molecular Cell Biology(2009) doi:10.1038/nrm2805.
- Like (3)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2026 MolecularCloud