Maximizing Biofuel Production in the Yeast R. toruloides through Metabolic Engineering
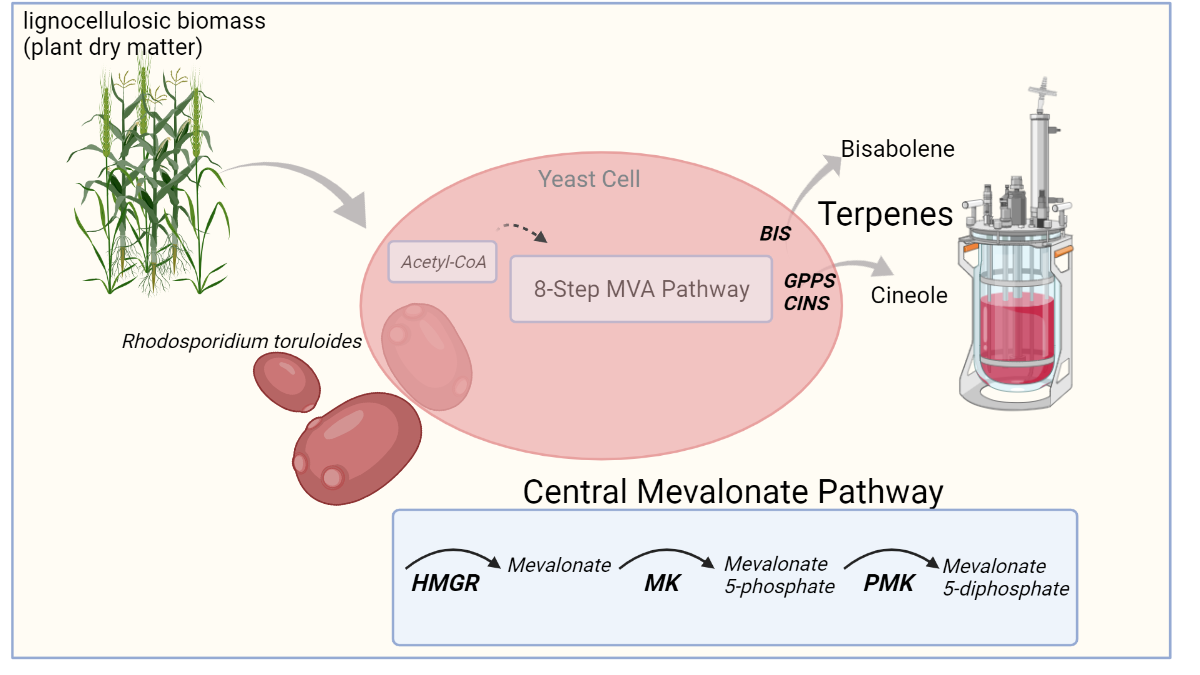
Lignocellulose or plant dry matter is an abundant renewable carbon resource that may be exploited as an energy source to reduce our dependency on fossil fuels. Lignocellulosic biomass consists primarily of cellulose, hemicellulose, and lignin, which microorganisms may process to support biofuel production. (Zoghlami et al. 2019). Nevertheless, lignocellulose's chemical and structural properties make this substrate resistant to degradation, resulting in low bioproduct yields, significantly hindering biofuel production. Although bacterial and yeast strains (e.g., Escherichia coli and Saccharomyces cerevisiae) have been engineered to improve lignocellulose processing, low yields and product-induced toxicity remain limiting factors to the use of these microbes. In contrast, Rhodosporidium toruloides has been identified as a promising microorganism for lignocellulose bioconversion to produce terpene-based biofuels (Yaegashi et al. 2017, Zhuang et al. 2019). Recent work by Kirby et al. 2021 shows the effectiveness of metabolic engineering through successive “Design-Build-Test-Learn” cycles supported by GenScript’s High-Throughput DNA Library Assembly service for systematic optimization of terpene production in R. toruloides.
Optimizing Terpene Production in R. toruloides
Metabolic engineering efforts had allowed Kirby and colleagues to develop R. toruloides strains with increased terpene yields, specifically α-bisabolene (GB2 strain) and 1,8-cineole (345 strain). For the GB2 strain, increased expression of Bisabolene synthase (BIS) achieved using two different promoters (i.e., PANT and PGAPDH) supported a 1.5 fold increase in α-bisabolene yield. Similarly, 1,8-cineole production was improved in the 345 strain by modifying the expression of GPP (GPPS) and 1,8-cineole (CINS) synthases. Therefore, by modifying the expression of terminal enzymes in the synthesis of α-bisabolene and 1,8-cineole, Kirby et al. were able to generate strains with improved capacity for terpene production.
Created with Biorender.com
R. toruloides is a carotenogenic red yeast adept at using various carbon sources such as those derived from lignocellulosic biomass. To optimize terpene yields, Kirby et al. 2021 targeted central enzymes in the mevalonate pathway for metabolic engineering to improve pathway flux in R. toruloides.
Recognizing that these newly developed R. toruloides strains could be further improved to achieve higher terpene yields, Kirby and colleagues focused on enzymes central to the mevalonate pathway (i.e., HMG-CoA reductase- HMGR, Mevalonate kinase- MK, and Phosphomevalonate kinase- PMK). Thus, Kirby et al. envisioned a mevalonate pathway engineering strategy using various mevalonate gene orthologs for HMGR, MK, and PMK each under the control of specific promoters (i.e., PANT, PSKP1, and PDUF) for expression in R. toruloides. Initially, orthologs were expressed individually in the optimized strains (i.e., GB2 and 345) and evaluated for their impact on terpene yields.
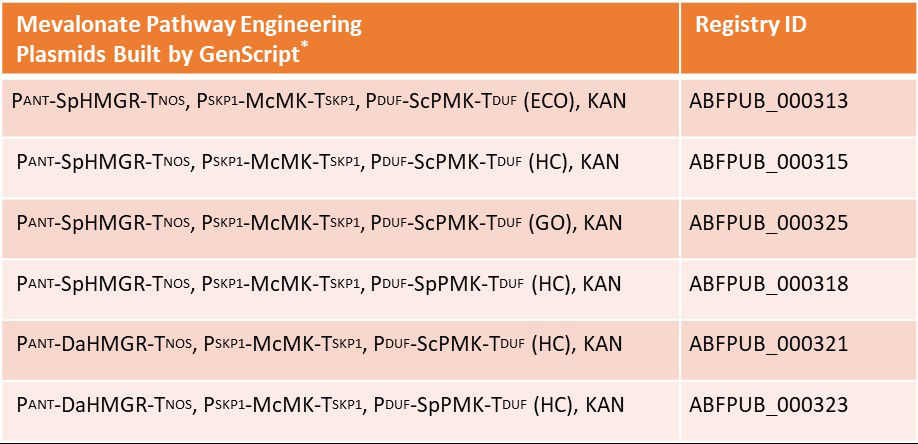
High throughput (HTP) DNA assembly provides the opportunity to expedite metabolic optimization efforts by accelerating the “Build” phase of the “Design-Build-Test-Learn” cycles. Therefore, Kirby and colleagues relied on GenScript’s HTP DNA assembly services to support central mevalonate enzyme gene synthesis and assembly into multiple plasmids. Based on their initial findings, specific orthologs were assembled combinatorially into plasmids for expression in either the GB2 or 345 strain.
Adapted from Kirby et al. 2021, https://creativecommons.org/licenses/by/4.0/
*HTP assembly was partly supported by an Innovation Grant sponsored by GenScript Biotech Corporation.
Silicibacter pomeroyi (SpHMGR), Delftia acidovorans (DaHMGR), Methanosaeta concilii (McMK), S. cerevisiae (ScPMK), Streptococcus pneumoniae (SpPMK). ECO, HC, and GO (GenScript’s codon optimization algorithm) correspond to the codon optimization method implemented.
Overall, Kirby and colleagues found that expression of two plasmids, PANT-SpHMGR, PSKP1-McMK, PDUF-SpPMK, and PANT-DaHMGR, PSKP1-McMK, PDUF-ScPMK in the 345 strain, significantly boosted 1,8-cineole production to above ~1.4 g/L. Although none of the constructs impacted α-bisabolene yield as substantially when expressed in the GB2 strain, the levels of these terpenes achieved by Kirby and colleagues represent the highest obtained to date from a microbe. Lastly, their findings provide a path to continue improving metabolic pathways to support higher terpene titers in R. toruloides.
Reference
- Like (3)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2024 MolecularCloud



